Introduction
“Survival analysis is a key technique in data-driven decision-making,
which is now central to public interest because of COVID-19. Applying
the correct technique for the specific question at hand is crucial for
credible public health inferences. If you are interested in assessing
how a risk factor or a potential treatment affects the progression of a
disease—such as how long a patient takes to recover—then survival
analysis techniques come into play. Survival analysis deeply respects
the ultimate source of its data, often the disease experience or even
the life and death of human patients. It seeks to exploit every last
drop of information that this experience can render for saving lives—in
particular, not only whether patients survived, but how long, and why.
And it strives to do so with minimal assumptions, so that the data are
truly driving the decision.”
—SAS Corporation
Setup of Data
. stset // show st setup of data
-> stset studytime, failure(died)
Survival-time data settings
Failure event: died!=0 & died<.
Observed time interval: (0, studytime]
Exit on or before: failure
──────────────────────────────────────────────────────────────────────────
48 total observations
0 exclusions
──────────────────────────────────────────────────────────────────────────
48 observations remaining, representing
31 failures in single-record/single-failure data
744 total analysis time at risk and under observation
At risk from t = 0
Earliest observed entry t = 0
Last observed exit t = 39
. describe // show variables in data
Contains data from https://www.stata-press.com/data/r18/drugtr.dta
Observations: 48 Patient survival in drug trial
Variables: 8 3 Mar 2022 02:12
───────────────────────────────────────────────────────────────────────────────────────
Variable Storage Display Value
name type format label Variable label
───────────────────────────────────────────────────────────────────────────────────────
studytime byte %8.0g Months to death or end of exp.
died byte %8.0g 1 if patient died
drug byte %8.0g Drug type (0=placebo)
age byte %8.0g Patient's age at start of exp.
_st byte %8.0g 1 if record is to be used; 0 otherwise
_d byte %8.0g 1 if failure; 0 if censored
_t byte %10.0g Analysis time when record ends
_t0 byte %10.0g Analysis time when record begins
───────────────────────────────────────────────────────────────────────────────────────
Sorted by:
Kaplan-Meier Survivor Function (per Gabriela Ortiz, Stata)
\[ S(t)=Pr(T>t) \]
. sts graph, scheme(michigan) // Kaplan-Meier Survivor Function
Failure _d: died
Analysis time _t: studytime
. graph export survival0.png, width(1000) replace
file
/Users/agrogan/Desktop/GitHub/newstuff/categorical/survival-analysis-and-event-hi
> story/survival0.png saved as PNG format
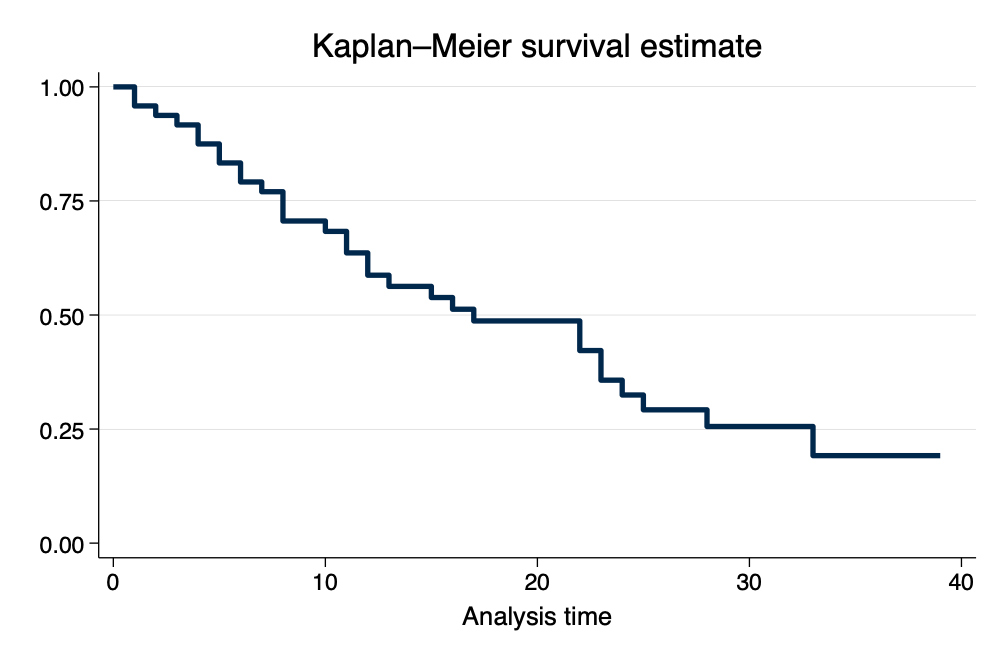
Kaplan-Meier Survivor Function
Graph Survival Curves
. stcurve, survival scheme(michigan) // survival curve
note: function evaluated at overall means of covariates.
. graph export survival1.png, width(1000) replace
file
/Users/agrogan/Desktop/GitHub/newstuff/categorical/survival-analysis-and-event-hi
> story/survival1.png saved as PNG format
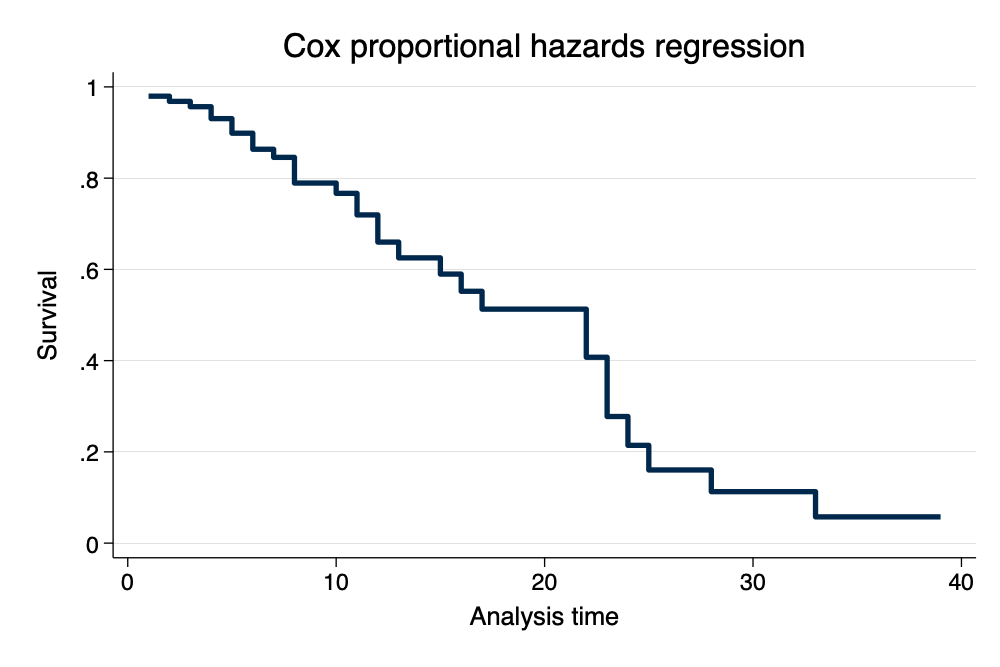
Survival Curve
. stcurve, survival at1(drug=0) at2(drug=1) scheme(michigan) // survival curve by group
note: function evaluated at specified values of selected covariates and overall means
of other covariates (if any).
. graph export survival2.png, width(1000) replace
file
/Users/agrogan/Desktop/GitHub/newstuff/categorical/survival-analysis-and-event-hi
> story/survival2.png saved as PNG format
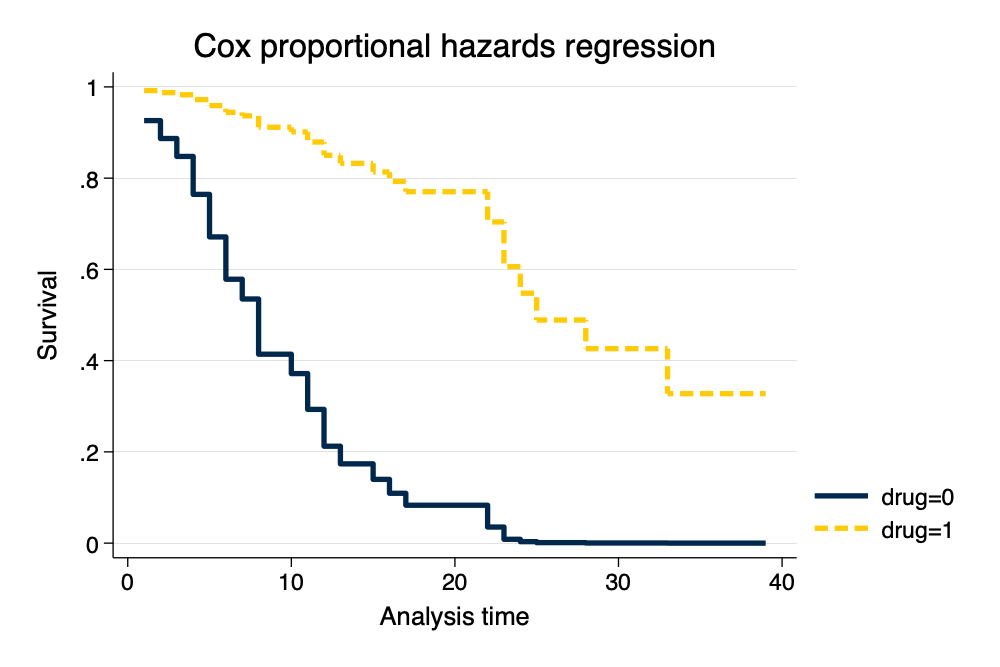
Survival Curve by Drug Group
Proportional Hazards Assumption
. estat phtest // formal test of PH assumption
Test of proportional-hazards assumption
Time function: Analysis time
─────────────┬──────────────────────────────────
│ chi2 df Prob>chi2
─────────────┼──────────────────────────────────
Global test │ 0.43 2 0.8064
─────────────┴──────────────────────────────────
. stphplot, by(drug) scheme(michigan) // graphical test of PH assumption
Failure _d: died
Analysis time _t: studytime
. graph export ph.png, width(1000) replace
file
/Users/agrogan/Desktop/GitHub/newstuff/categorical/survival-analysis-and-event-hi
> story/ph.png saved as PNG format
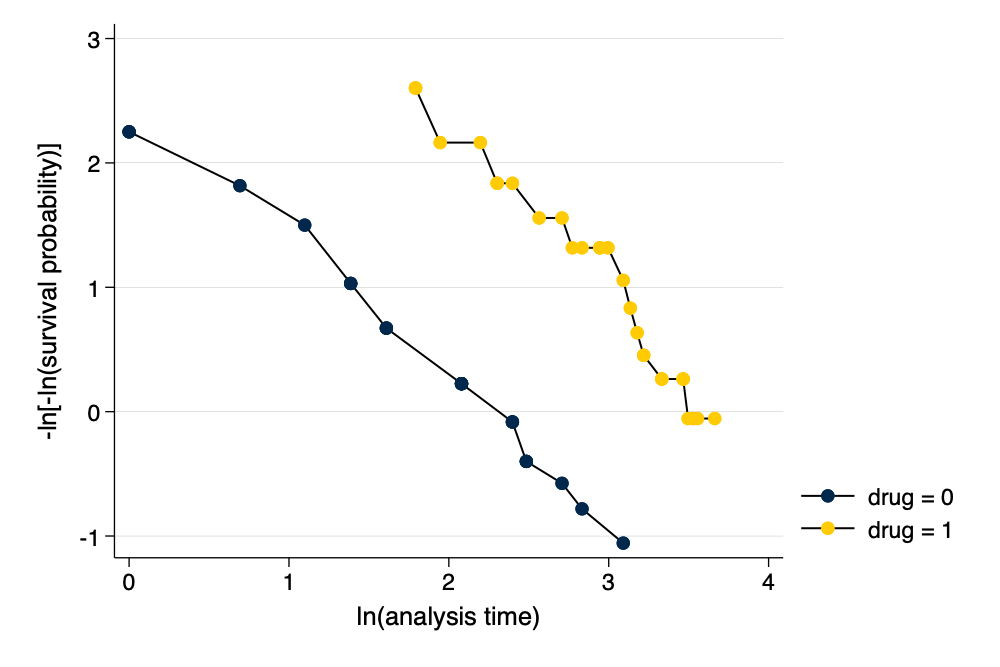
Graphical Assessment of Proportional Hazards
Assumptions
References
Johnson, L. L., & Shih, J. H. (2007). CHAPTER 20 - An
Introduction to Survival Analysis (J. I. Gallin & F. P. Ognibene,
eds.).
https://doi.org/https://doi.org/10.1016/B978-012369440-9/50024-4
Ragnar Frisch Centre for Economic Research (2020). Event History
Analysis, Survival Analysis, Duration Analysis ,Transition Data
Analysis, Hazard Rate Analysis. Oslo, Norway.



